Introduction
The market for therapeutic antibodies has dramatically expanded over the past decades since their first approval in 1986. Antibodies, which are proteins, form their structure and exert their activity through a complex series of non-covalent bonds and may lose their activity due to various external stimuli. Therefore, the evaluation of structural stability is extremely important in the development and formulation of candidate antibodies. Thermodynamic stability of antibodies is generally evaluated by DSC (differential scanning calorimetry) and circular dichroism (CD) spectroscopy.1, 2) CD spectroscopy is an easy and rapid method for obtaining information on the secondary and tertiary structure of proteins in solution and can be used to directly evaluate the protein structural change caused by heat. Recently, Micsonai et al. developed the BeStSel algorithm that can accurately estimate the secondary structure composition from the CD spectrum by taking into account the parallel-antiparallel orientation of the β-strands and the twist of the antiparallel β-sheets. BeStSel has the following features: 1) high estimation accuracy for a wide range of proteins, including β-structure-rich-proteins such as antibodies, 2) providing eight types of secondary structure information 3) a capability to predict the protein fold following the CATH classification, and 4) an open web server.3-7) While many academic researchers use the BeStSel web server, researchers in biopharma who need to work in a GxP environment have not been able to benefit from BeStSel. To make BeStSel accessible to biopharma, we developed Spectra Manager™ Ver.2.5 CFR BeStSel as an add-in software for Spectra Manager™, a control and analysis platform for CD spectrometers, which is compatible with GxP. Here, we report the detailed tracking of heat stress-induced conformational changes of human Immunoglobulin G (h-IgG) and Herceptin® using Spectra Manager™ Ver.2.5 CFR BeStSel.
Experimental
Measurement system

Materials
•Herceptin®(trastuzumab, Roche) powder dissolved in ultrapure water to 0.025 mg/mL
•IgG, from human serum (Sigma Aldrich) powder dissolved in ultrapure water to 0.025 mg/mL
Methods
Table 1. Measurement conditions for far-UV/CD
| Parameter | |
| Bandwidth | 1.0 nm |
| D.I.T. | 2 sec |
| Data pitch | 0.1 nm |
| Scan speed | 50 nm/min |
| Accumulations | 4 |
| Pathlength | 5 mm |
| Temp. range | 30 – 85 degree C |
Results
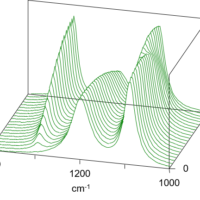
The thermal denaturation spectra of Herceptin® and h-IgG are shown (Fig. 1a, 1b). Both antibodies showed a spectral shape with a minimum around 217 nm, which is characteristic of the β-sheet at 30 degree C, and the peak intensity decreased as the temperature increased.
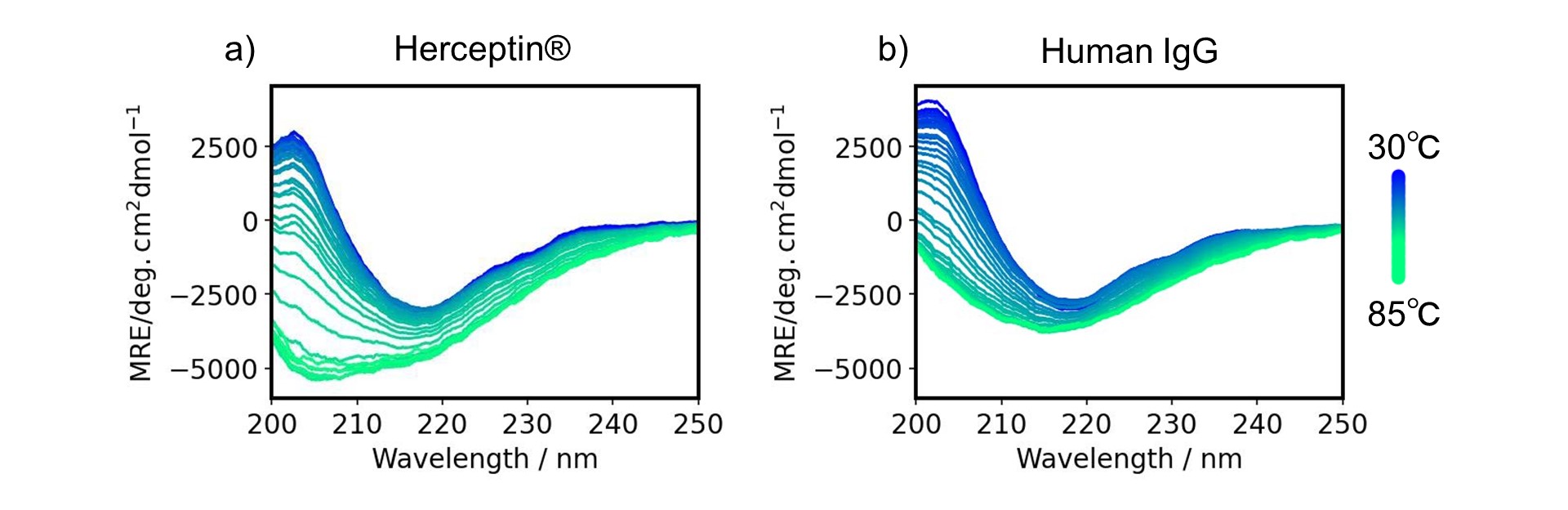
Figure 1-1. Thermal denaturation spectra for a) Herceptin®, and b) human IgG
In order to analyze these structural changes in detail, secondary structure estimation was performed on the spectra as a function of temperature using Spectra Manager™ Ver.2.5 CFR BeStSel to determine the change in the ratio of each secondary structure with temperature. The secondary structure ratio of β-strands and “others” including random coils at room temperature was consistent with that for human IgG (PDB ID 1IGT) obtained from the X-ray crystal structure (data not shown). The ratio of “others” increased while the ratio of β-strand decreased with heating for both Herceptin® and human IgG (Fig. 1c, 1d). The Tm values for Herceptin® and human IgG estimated from these curves were consistent with previously reported values.8, 9)
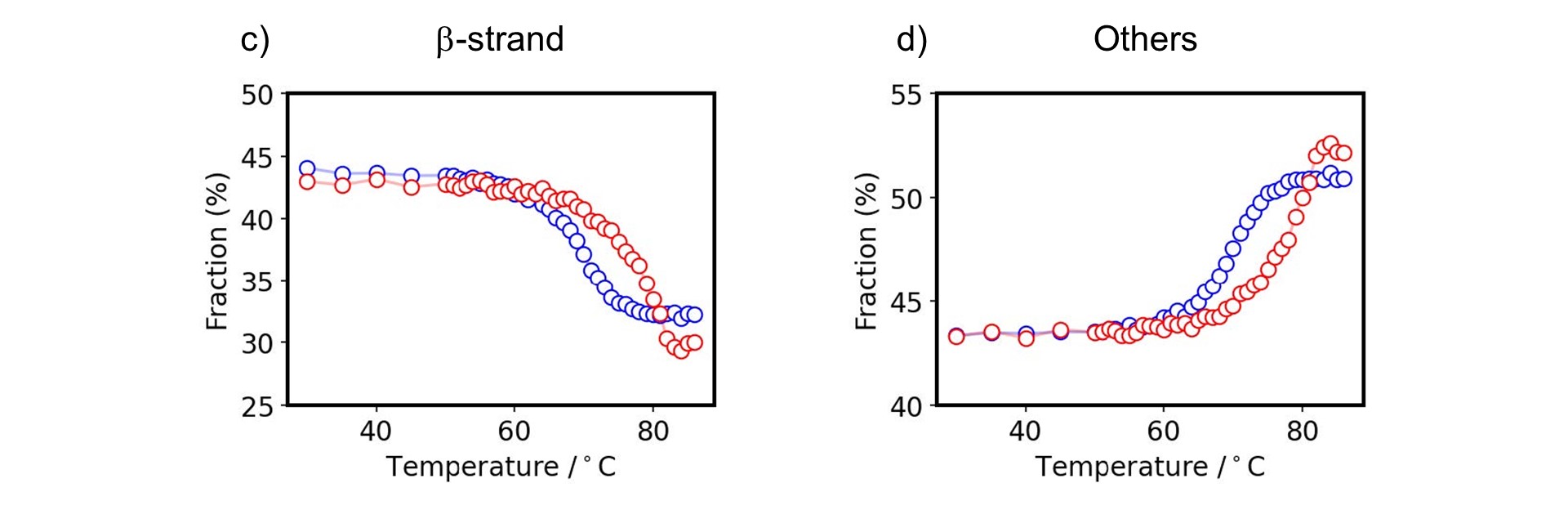
Figure 1-2. Melting property of antibodies
c) β-strand, d) others fraction of Herceptin® (red) and human IgG (blue)
The advantage of BeStSel is not only its high accuracy in analyzing β-structure-rich-proteins, but also its feature of being able to calculate the fraction of eight secondary structure elements, including three types of twisting in antiparallel β-sheets (Fig. 2).
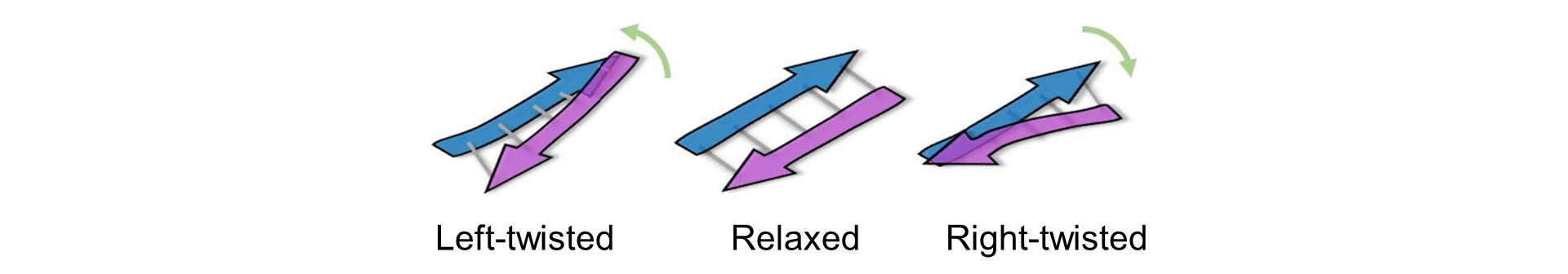
Figure 2. Twisting of β-strand
Fig. 3 shows the detailed secondary structure change as a function of temperature.
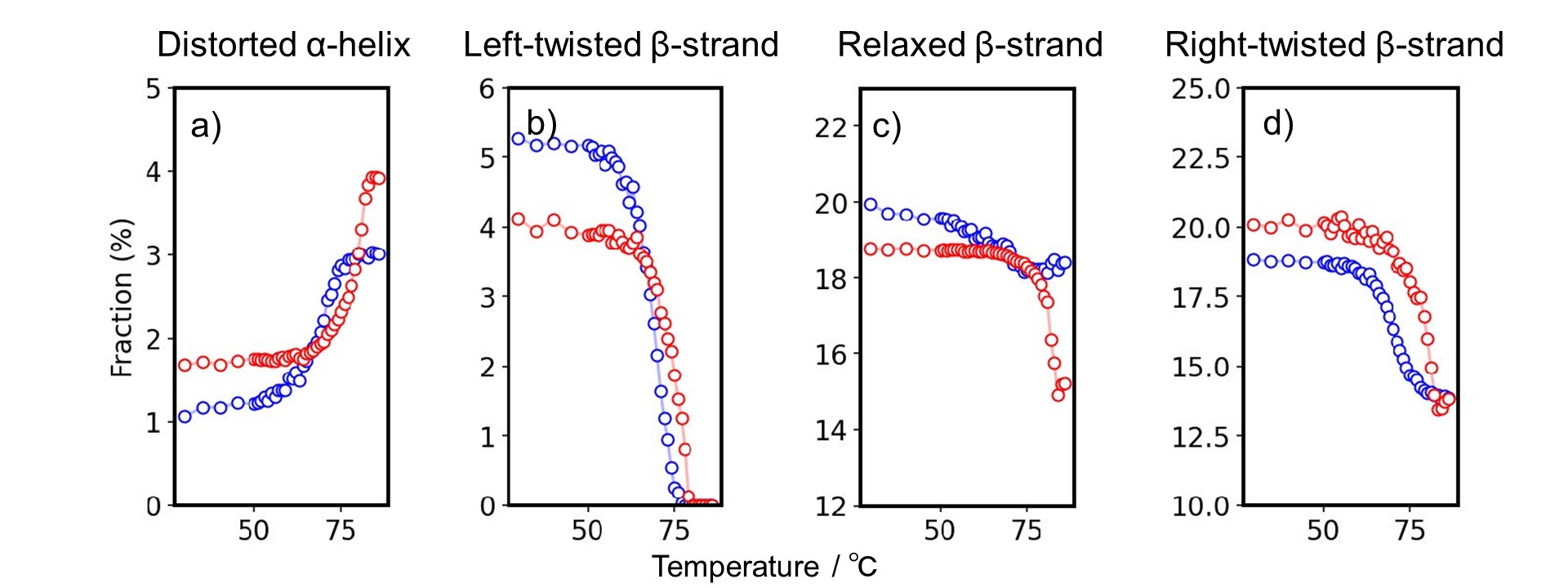
Figure 3. Detailed secondary structure changes as a function of temperature.
Estimated secondary structure fraction of a) distorted α-helix, b) left-twisted, c) relaxed, and d) right-twisted antiparallel β-strand of Herceptin® (red) and human IgG (blue).
There is a significant difference between Herceptin® and h-IgG in changes in the distorted α-helix, left-twisted β-strand, relaxed β-strand and right-twisted β-strand. At 30 degree C, h-IgG and Herceptin have a similar secondary structure, but it was found that they have different denaturation mechanisms by using BeStSel. BeStSel is shown to be useful in understanding the detailed mechanisms of structure formation in proteins, including antibody drugs.
In addition, Spectra Manager™ Ver.2.5 CFR BeStSel allows this detailed protein structural analysis to work in GxP compliant environment.
Conclusion
- Spectra Manager™ Ver.2.5 CFR BeStSel enables the tracking of detailed structural changes of antibody drugs.
- Spectra Manager™ Ver.2.5 CFR BeStSel satisfy the GxP compliant environment and offer CSV, ER/ES, and DI for practice ALCOA+.
References
Poster Session at 14th annual PEGS Europe (Protein & Antibody Engineering Summit, November 14 – 16, 2022, in Barcelona, Spain)
Satoko Suzuki1, Taiji Oyama1, Ai Yamane1, Yasuo Horiguchi1, András Micsonai2, József Kardos2, Ken-ichi Akao1
1JASCO Corporation, Hachioji, Tokyo, 192-8537, Japan, 2ELTE NAP Neuroimmunology Research Group, Department of Biochemistry, Institute of Biology, ELTE Eötvös Loránd University, Budapest H-1117, Hungary
1) Ama, D., Hasegawa, J., Uchiyama, S., and Fukui, K. (2010). Netsu sokutei (Japan), 38, 9-15.
2) Greenfield, N. J. (2006). Nat. Protoc, 1, 2733-2741.
3) Micsonai, A., Wien, F., Kernya, L., Lee, Y. H., Goto, Y., Réfrégiers, M., & Kardos, J. (2015). Proc. Natl. Acad. Sci. U.S.A., 112, E3095–E3103.
4) Micsonai, A., Wien, F., Bulyáki, É., Kun, J., Moussong, É., Lee, Y. H., Goto, Y., Réfrégiers, M., & Kardos, J. (2018). Nucleic Acids Res., 46, W315-W322.
5) Micsonai, A., Bulyáki, É., & Kardos, J. (2021). Methods in molecular biology (Clifton, N.J.), 2199, 175–189.
6) Micsonai, A., Moussong, É., Lee, Y. H., Murvai, N., Tantos, Á., Tőke, O., Réfrégiers, M., Wien, F., & Kardos, J. (2022). Front. mol. biosci, 9
7) Micsonai, A., Moussong, É., Wien, F., Boros, E., Vadászi, H., Murvai, N., Lee, Y. H., Molnár, T., Réfrégiers, M., Goto, Y., Tantos, Á., & Kardos, J. (2022). Nucleic Acids Res., 50, W90-W98.
8) Moro Pérez, L., Rodríguez Taño, A. C., Martín Márquez, L. R., Gómez Pérez, J. A., Valle Garay, A., & Blanco Santana, R. (2019). PloS one, 14, e0215442.
9) Moggridge, J., Biggar, K., Dawson, N., & Storey, K. B. (2017). Technology in cancer research & treatment, 16, 997–1005.