Introduction
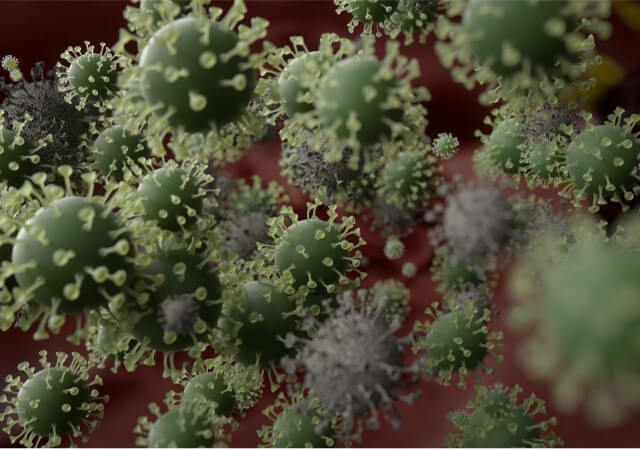
The coronavirus S glycoproteins sit on the surface of the virus particle (Figure 1) and facilitates receptor binding and membrane fusion which leads to entry of the virus into host cells. Therefore, the S proteins are the main target for neutralizing binding and therapeutic drug design.
VHH antibodies (variable domain of heavy chain of heavy chain antibodies) are small, single domain antibodies corresponding to the variable region of a heavy chain camelid antibody (Figure 1). They are highly stable with respect to heat, have high affinity and binding specificity, a low molecular weight which increases mass production efficiency, and can be easily modified. Since the structure of antibody drugs may change and cause a lose in their activity depending on the formulation conditions and storage environment, it is necessary to evaluate their stability in order to ensure quality. In particular, thermal denaturation is highly reversible depending on the protein, increasing the need to determine denaturation temperature and refolding ability in the stability evaluation of antibody drugs.
In this note, we evaluated the thermal stability and refolding ability of 3 different types of VHH antibodies using a J-1500 CD spectrometer. The secondary structure percentages were estimated using the BeStSel program to further correlate the denaturation temperatures with structural changes to VHH antibody framework.
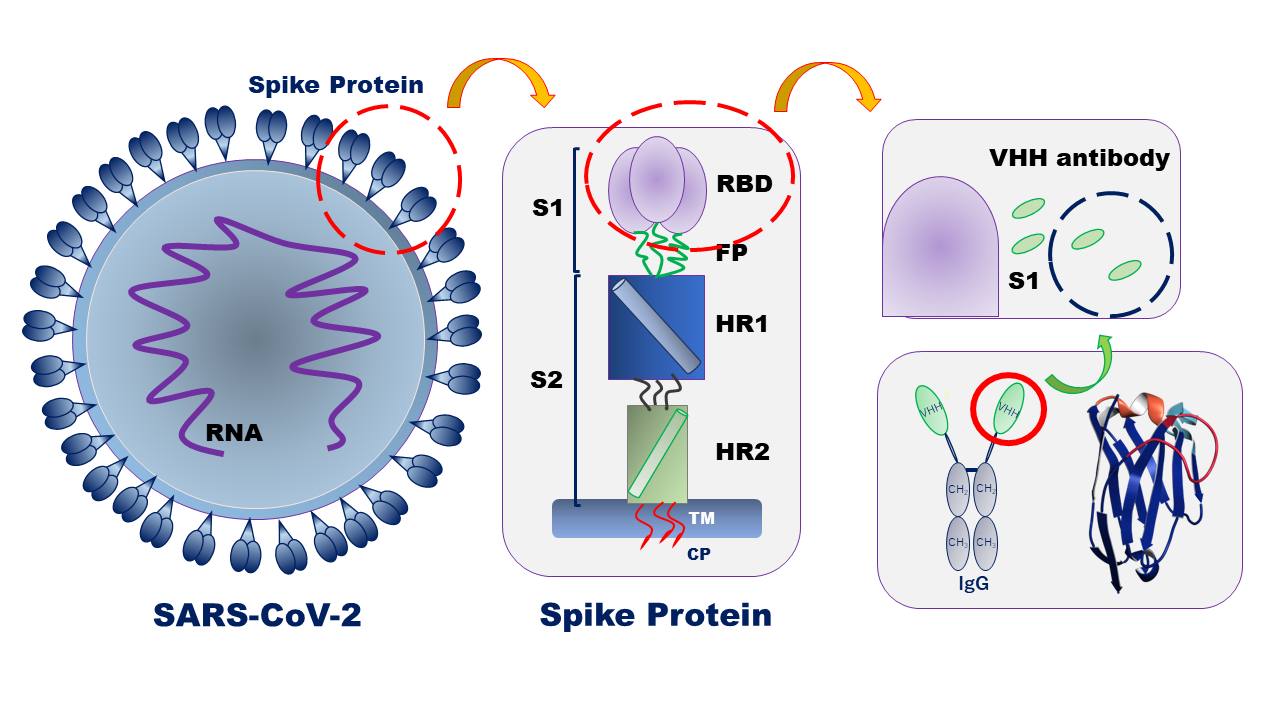
Figure 1. Coronavirus molecules image (left) and schematics of the spike proteins and VHH antibody structures.

BeStSel (http://bestsel.elte.hu/) is a secondary structure analysis program developed by the research group of Dr. József Kardos at ELTE Eötvös Loránd University, Budapest1, 2). The algorithm deconvolutes β-sheet structure into four substructures, taking into account the twist between β-strands. This enables highly accurate secondary structure analysis of β-sheet-rich proteins like protein aggregates such as amyloid fibrils and IgG. Therefore, BeStSel was used in this study for the secondary structure analysis of VHH antibodies.
Experimental
Antibody denaturation study
The denaturation curve of the VHH antibody was obtained by heating the sample temperature from 25 to 90ºC while measuring the CD value at 217 nm, indicative of β-sheet structure. The starting temperature, onset temperature (To), denaturation temperature (Tm), and end temperature (Te) were calculated from the obtained denaturation curves.
<Sample>
Anti-SARS-CoV-2 VHH antibody (3 kinds) : No.1 (0.4 mg/mL), No.2 (0.28 mg/mL), No.3 (0.2 mg/mL)
Buffer : 20 mM PBS
<Measurement parameters>
Wavelength : 217 nm
Bandwidth : 1 nm
D.I.T.* : 2 sec.
Temp. interval : 1ºC
Temperature gradient : 1ºC /min.
Temperature range : 25 – 90ºC
Secondary structure changes due to refolding
The CD value at 217 nm was measured from 25ºC to the end temperature (Te) obtained from the denaturation curves, and then cooled back to 25ºC . CD spectra were measured at the start temperature, onset (To), denaturation temperature (Tm), and end temperature (Te). Secondary structure analysis was performed on the obtained CD spectra using BeStSel and the refolding ability was calculated from the amount of change in the β-sheet structure at 25ºC.
<Samples>
Anti-SARS-CoV-2 VHH antibody (3 kinds) : No.1 (0.1 mg/mL), No.2 (0.1 mg/mL), No.3 (0.1 mg/mL)
Buffer : 20 mM PBS
<Measurement parameters>
Wavelength : 217 nm
Bandwidth : 1 nm
D.I.T.* : 4 sec.
Temp. interval : 1ºC
Temperature gradient : 1ºC/min.
Temperature range : 25 – 75ºC (No. 1, 2), 25 – 80ºC (No. 3)
Spectra Measurement
<Measurement parameters>
Wavelength : 250 – 200 nm
Bandwidth : 1 nm
D.I.T.* : 4 sec.
Scanning speed : 50 nm/min.
Data interval : 0.1 nm
Accumulations : 9 times
*D.I.T.: Data Integration Time

J-1500 CD Spectrometer with MPTC-513 Multi-Position Peltier Cell Changer
Keywords
SARS-CoV-2, Coronavirus, Antibody drugs, VHH, Refolding, Onset, Denaturation temperature, Secondary structure, BeStSel, Circular Dichroism
Results
Antibody denaturation study
Figure 2 shows the denaturation curve of the CD at 217 nm which reflects changes in β-sheet structure. Table 1 shows the onset (To), denaturation temperature (Tm), and end temperature (Te) obtained from the denaturation curve. Comparing the denaturation temperatures, it can be seen that the thermal stability of the samples decreasing in the following order: Sample 3, 1, and 2.
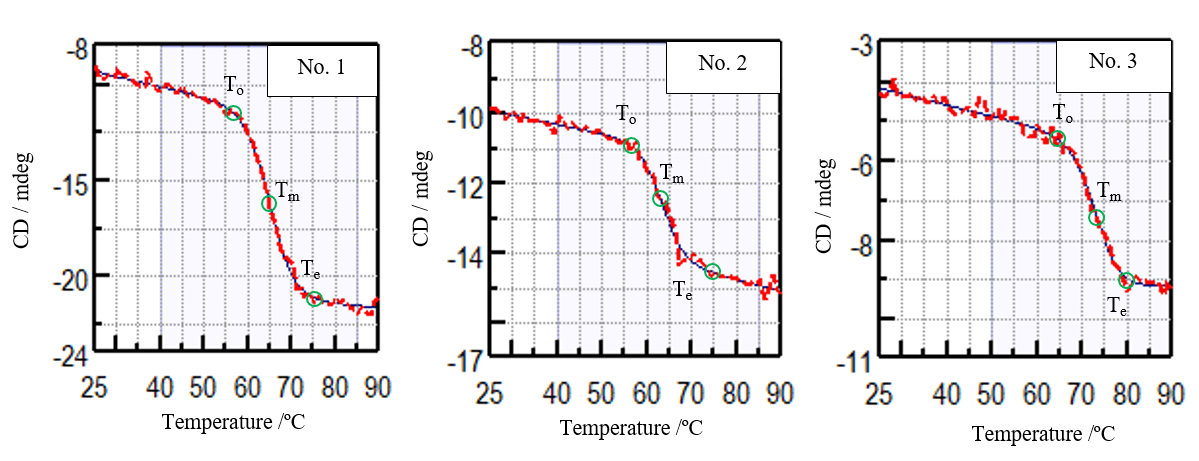
Figure 2. Measured denaturation curves of VHH antibodies at 217 nm (red) and their fitted curves (blue).
Table 1. Analysis results of onset temperature (To), denaturation temperature (Tm), and end temperature (Te).
| Samples | To /ºC | Tm /ºC | Te /ºC |
|---|---|---|---|
| No. 1 | 57 | 65 | 75 |
| No. 2 | 57 | 63 | 75 |
| No. 3 | 65 | 73 | 80 |
Secondary structure changes due to refolding
In order to reduce the effect of aggregation, the sample was heated to the end temperature (Te) determined from the previously obtained denaturation curves and cooled immediately after the sample denatured. Figure 3 shows the denaturation and refolding curves at 217 nm and the CD spectra at the samples’ start temperature, onset temperature (To), denaturation temperature (Tm), and end temperature (Te).
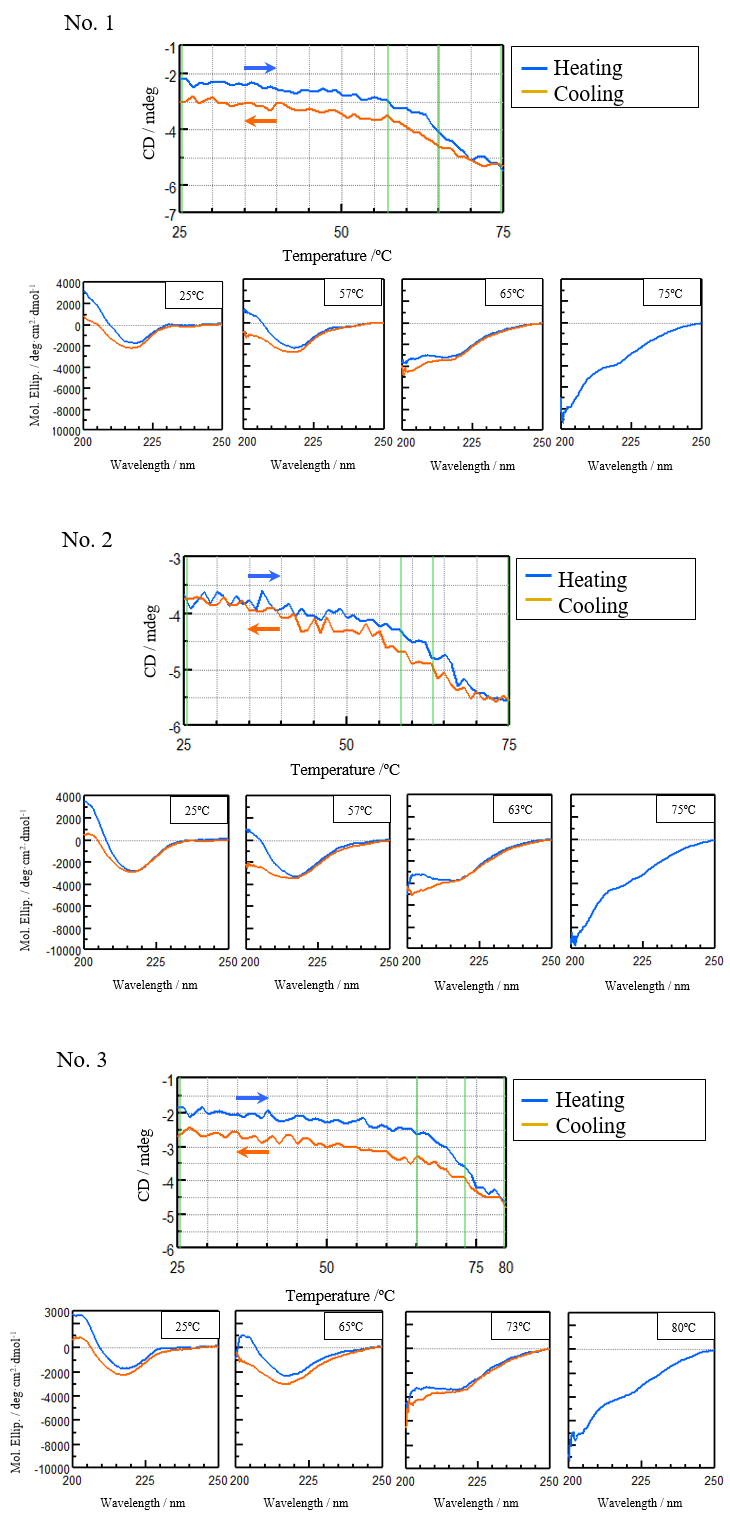
Figure 3. Denaturation curves (top) and CD spectra (bottom) of VHH antibodies at their start temperatures, To, Tm, and Te (bottom).
Table 2 and Figure 4 show the result of the secondary structure analysis by using BeStSel.
Table 2. Secondary structure analysis results calculated using the BeStSel program.
No.1
| Temp/ºC | α-Helical/% | β-sheet/% | Turn/% | Others*/% |
|---|---|---|---|---|
| 25 | 1.5 | 41.3 | 12.5 | 44.6 |
| 57 | 1.9 | 39.1 | 13.0 | 46.0 |
| 65 | 3.0 | 32.3 | 14.5 | 50.2 |
| 75 | 5.0 | 26.3 | 15.6 | 53.1 |
| 65 | 3.4 | 30.6 | 14.7 | 51.4 |
| 57 | 2.4 | 36.4 | 12.5 | 45.3 |
| 25 | 2.0 | 40.2 | 12.5 | 45.3 |
No.2
| Temp/ºC | α-Helical/% | β-sheet/% | Turn/% | Others*/% |
|---|---|---|---|---|
| 25 | 2.2 | 43.6 | 11.6 | 42.7 |
| 57 | 2.3 | 39.8 | 12.4 | 45.4 |
| 63 | 3.2 | 32.6 | 14.8 | 49.4 |
| 75 | 2.6 | 25.6 | 17.0 | 54.8 |
| 63 | 3.7 | 31.2 | 14.5 | 50.6 |
| 57 | 3.4 | 34.8 | 13.5 | 48.4 |
| 25 | 2.4 | 41.4 | 12.4 | 44.1 |
No.3
| Temp/ºC | α-Helical/% | β-sheet/% | Turn/% | Others*/% |
|---|---|---|---|---|
| 25 | 1.7 | 42.9 | 12.7 | 42.6 |
| 65 | 1.7 | 39.6 | 14.2 | 44.4 |
| 73 | 3.2 | 29.4 | 15.6 | 51.8 |
| 80 | 2.7 | 26.4 | 17.1 | 53.8 |
| 73 | 2.8 | 28.8 | 16.2 | 52.2 |
| 65 | 2.7 | 34.7 | 13.6 | 49.0 |
| 25 | 2.4 | 40.1 | 13.2 | 44.2 |
*3,10-helix, π-helix, β-bridge, bend, loop/irregular, invisible regions of the structure
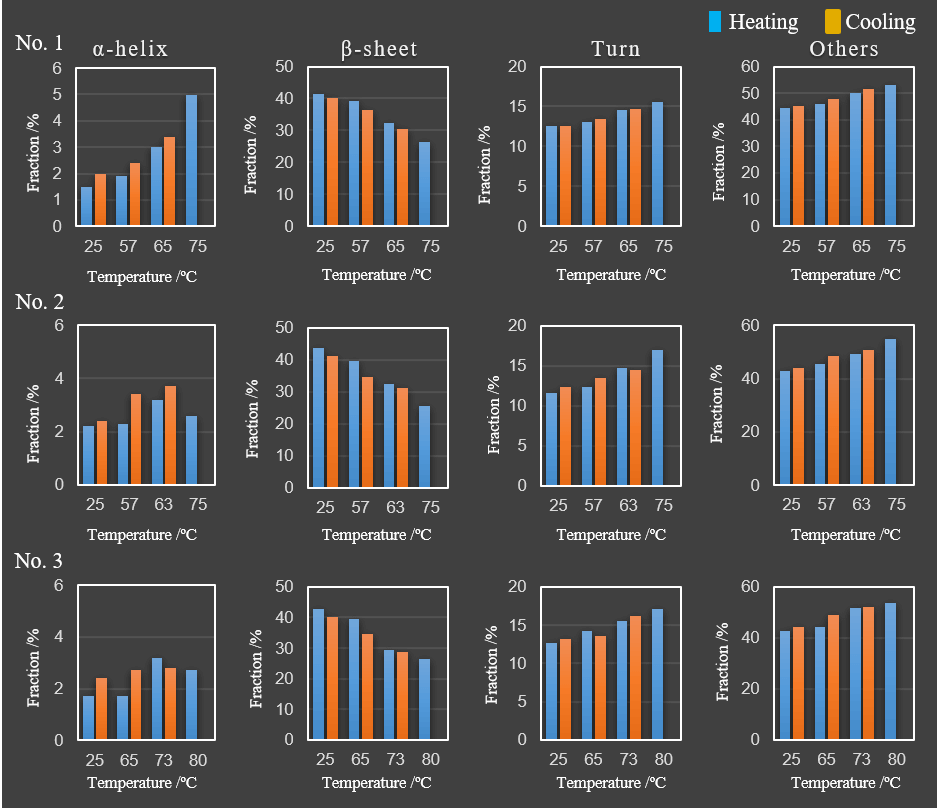
Figure 4. Changes in secondary structure percentages as a function of temperature for the three antibody samples.
Conclusion
The results show that β-sheet structures decrease and turn and other structures increase with increasing temperatures for all samples. In addition, the α-helix percentage increased with increasing temperature for sample No. 1 only. The thermal reversibility study indicates that the β-sheet, turn, and other structure of all three samples are reversible, as well as the α-helix structure of No. 1 sample. While the framework of VHH antibodies are considered conserved, the structure of CDR (complementarity determining regions) that bind to the antigens is variable. The framework is primarily composed of β-sheet structure and any changes in structure can potentially affect the structure of the CDR. Focusing on the β-sheet secondary structure percentages, the effective refolding before and after heating were calculated for samples No. 1, No. 2 and No. 3 and were 97.3%, 95.0% 93.5%, respectively. Therefore the ability of the samples to refold after heating and cooling assume the following order, No. 1, No. 2 and No. 3.
References
1) A. Micsonai, F. Wien, L. Kernya, Y.H. Lee, Y. Goto, M. Refregiers, J. Kardos, Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy, PNAS, 112 (2015) 3095-3103.
2) A. Micsonai, F. Wien, E. Bulyaki, J. Kun, E. Moussong, Y.H. Lee, Y. Goto, M. Refregiers, J. Kardos, BeStSel: a web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra, Nucleic Acids Res, 46 (2018) 315-322.
Acknowledgements
Special thanks to RePHAGEN for providing VHH antibody samples
Many thanks to Dr. József Kardos at ELTE Eötvös Loránd University for scientific advice