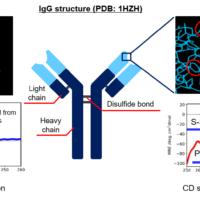
Introduction
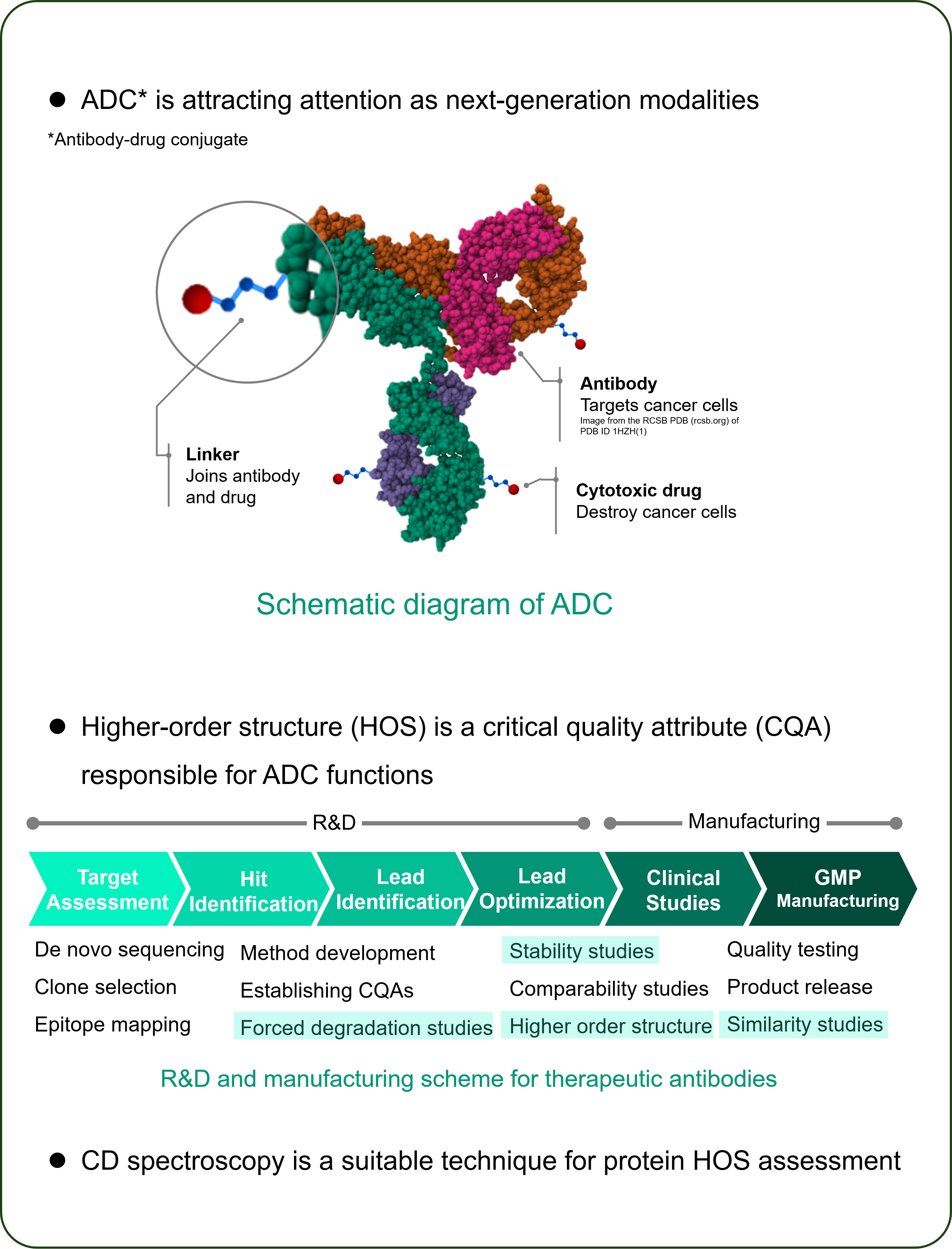
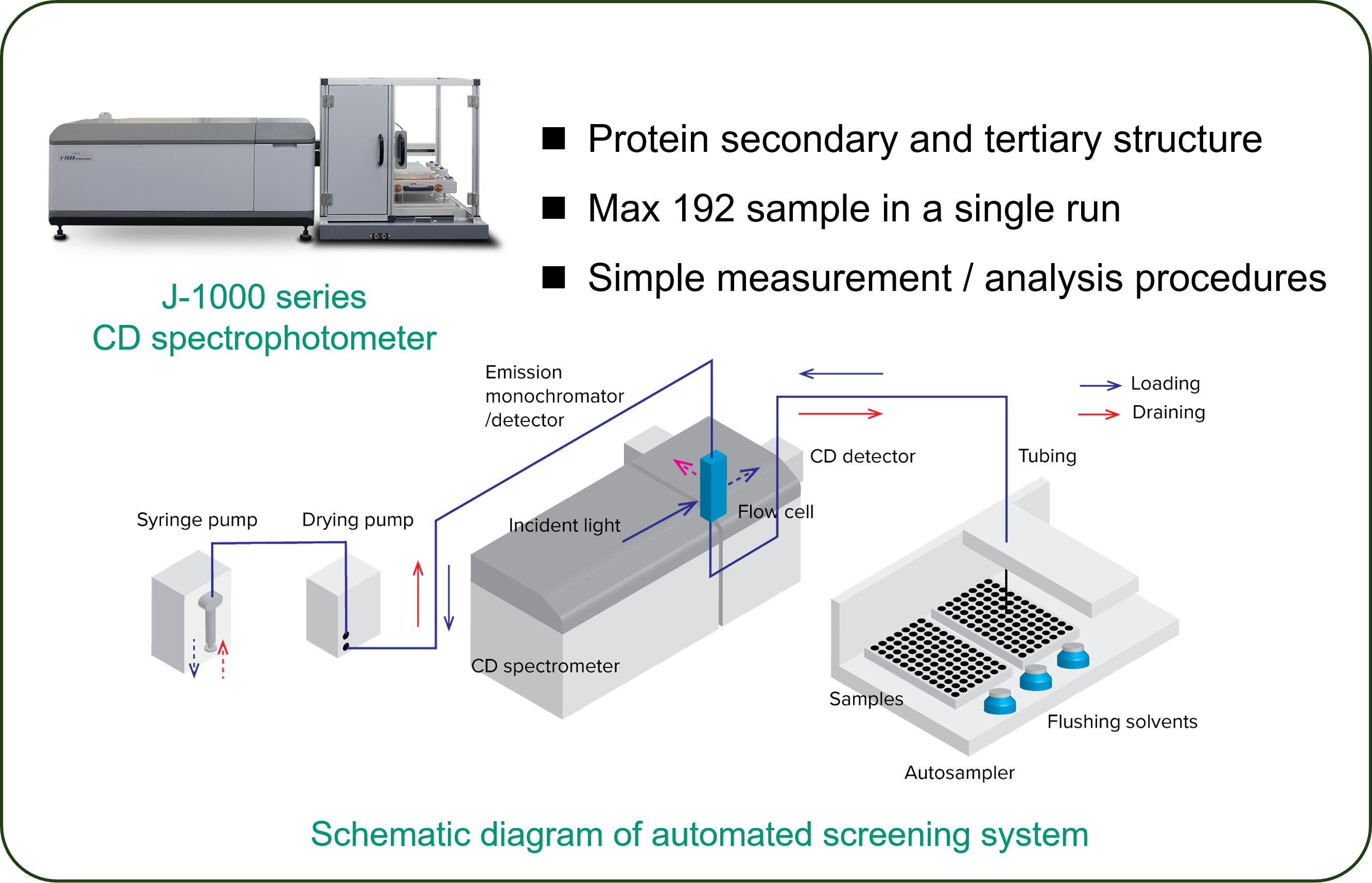
Experimental
Sample
- Kadcyla®
- lEnhertu®
-HER2-targeted ADC consisting of trastuzumab and cytotoxic drug - Herceptin® (trastuzumab)
– HER2-targeted antibody therapeuticHerceptin and Kadcyla are registered trademarks of Genentech inc.
Enhertu is a registered trademark of Daiichi Sankyo.Instrument
Results
pH STABILITY
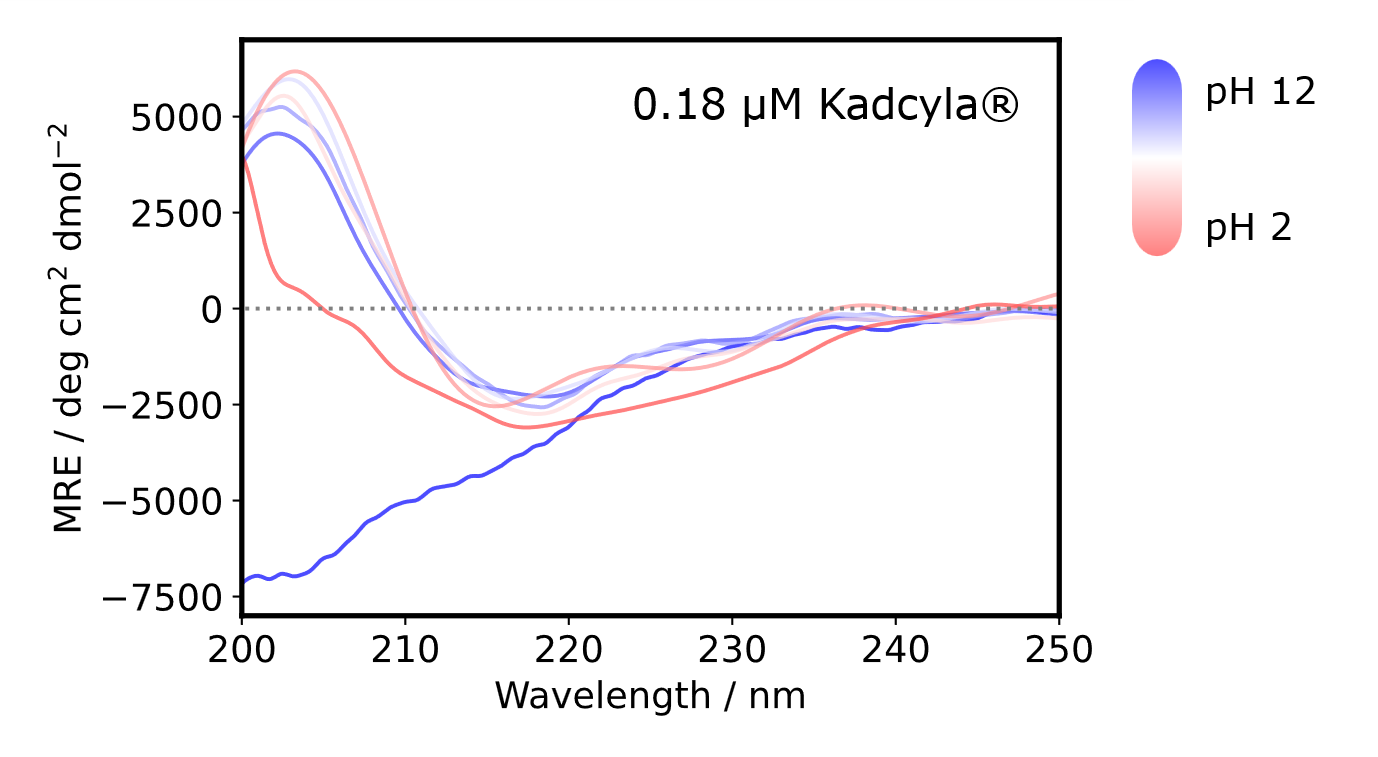
CD spectra were obtained at pH 2, 3, 5, 7, 10, 11, and 12 and converted into MRE (molar residue ellipticity) spectra for normalization.
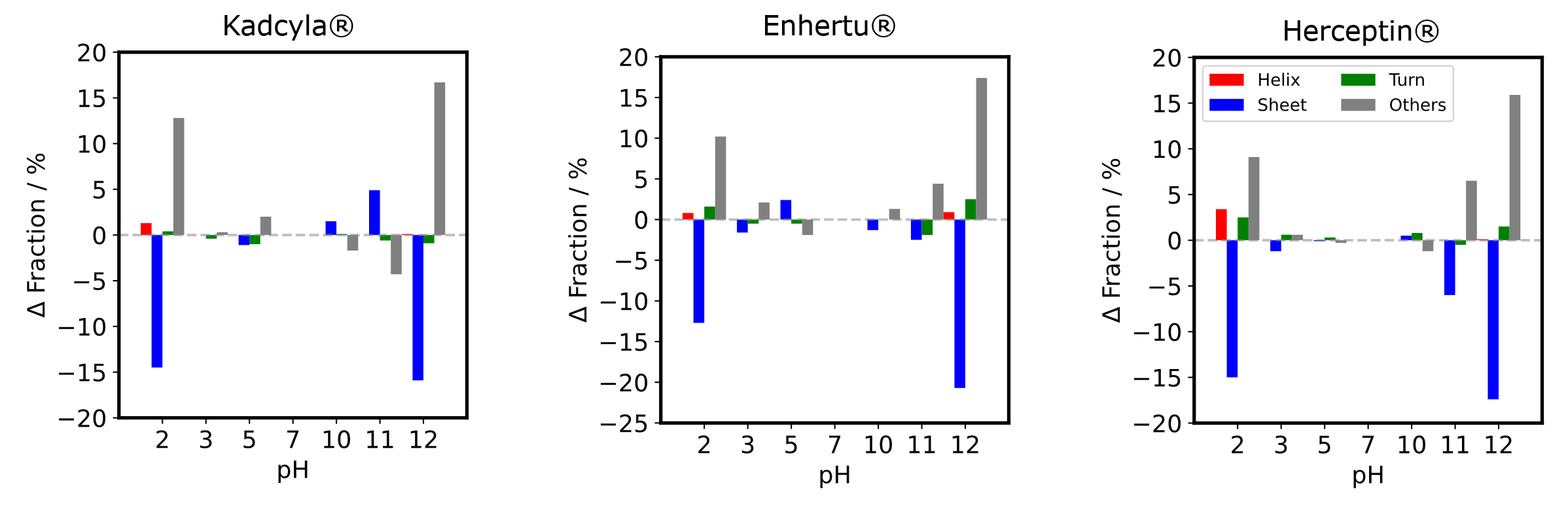
- Secondary structure compositions were estimated from the MRE spectra using BeStSel, a high-performance secondary structure determination algorithm developed by Dr. József Kardos and Dr. András Micsonai (https://bestsel.elte.hu)(2-6).
- The difference in fraction between pH 7 and each pH was plotted. Between pH 3 and pH 10, each structure composition seems to remain, however, strand and others compositions shows a significant change at pH 2, 11, and 12.
HEAT STABILITY
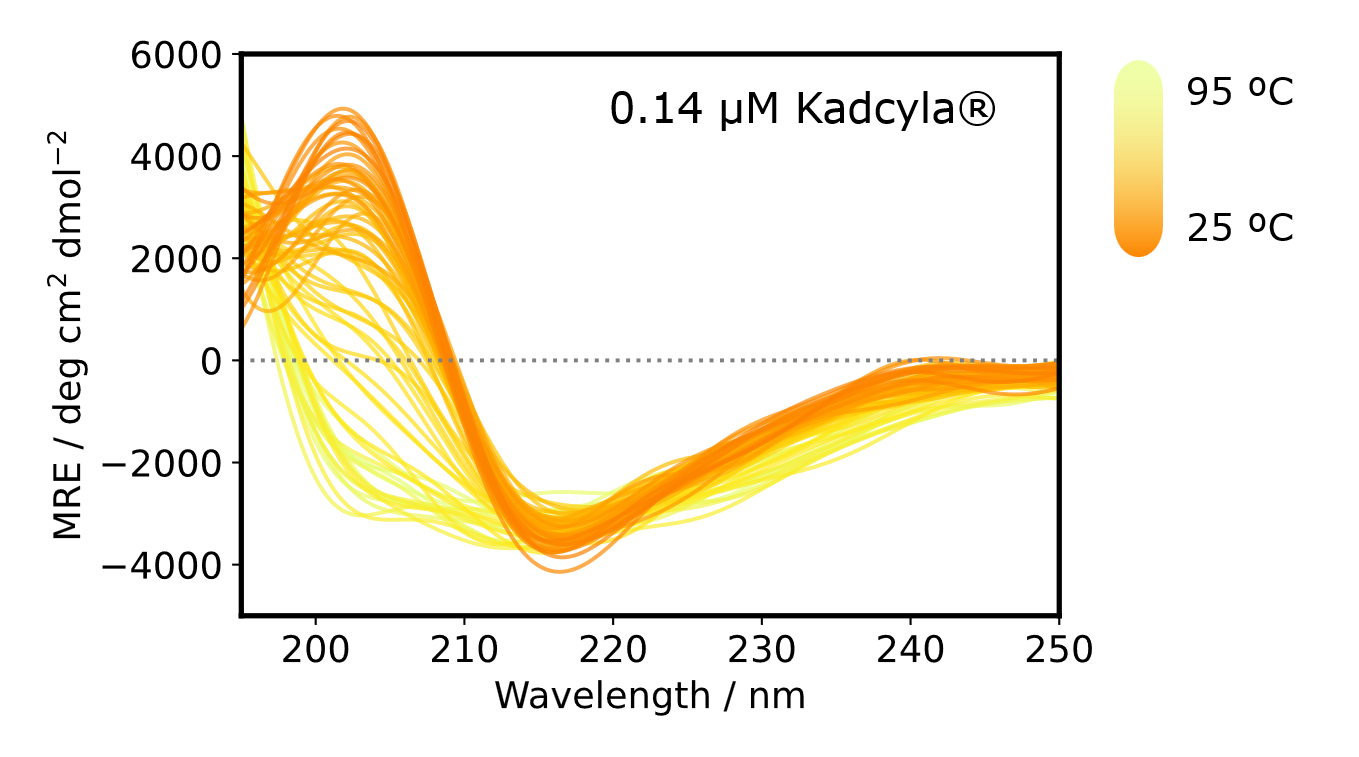
CD spectra were obtained while changing the temperature from 25 ºC to 95 ºC by 1 ºC interval and converted to MRE spectra for normalization.
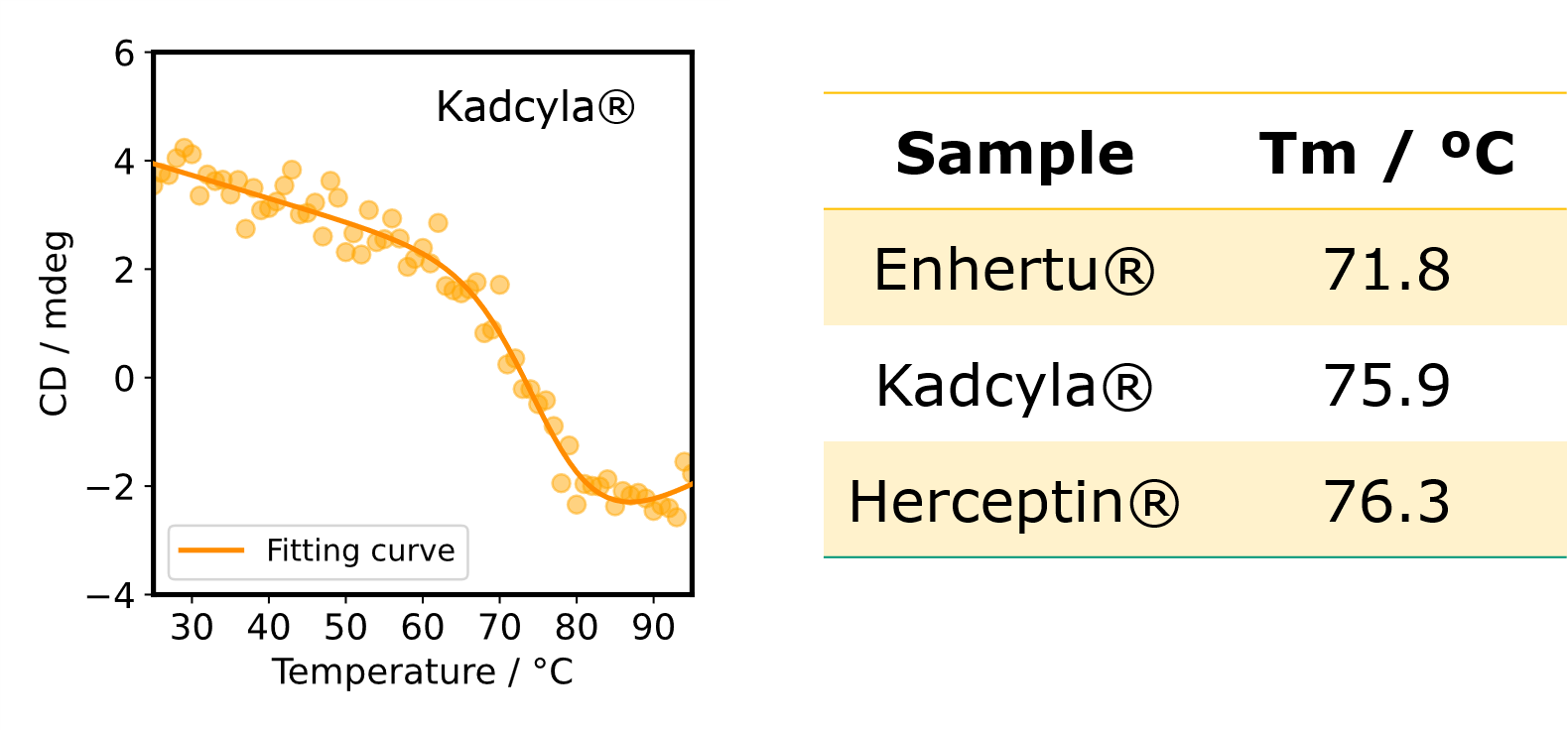
Melting temperature (Tm) was estimated from the denaturation curve to check the thermal stability. The denaturation curve was obtained by plotting the CD signal at 203 nm which is characteristic of β-strand.
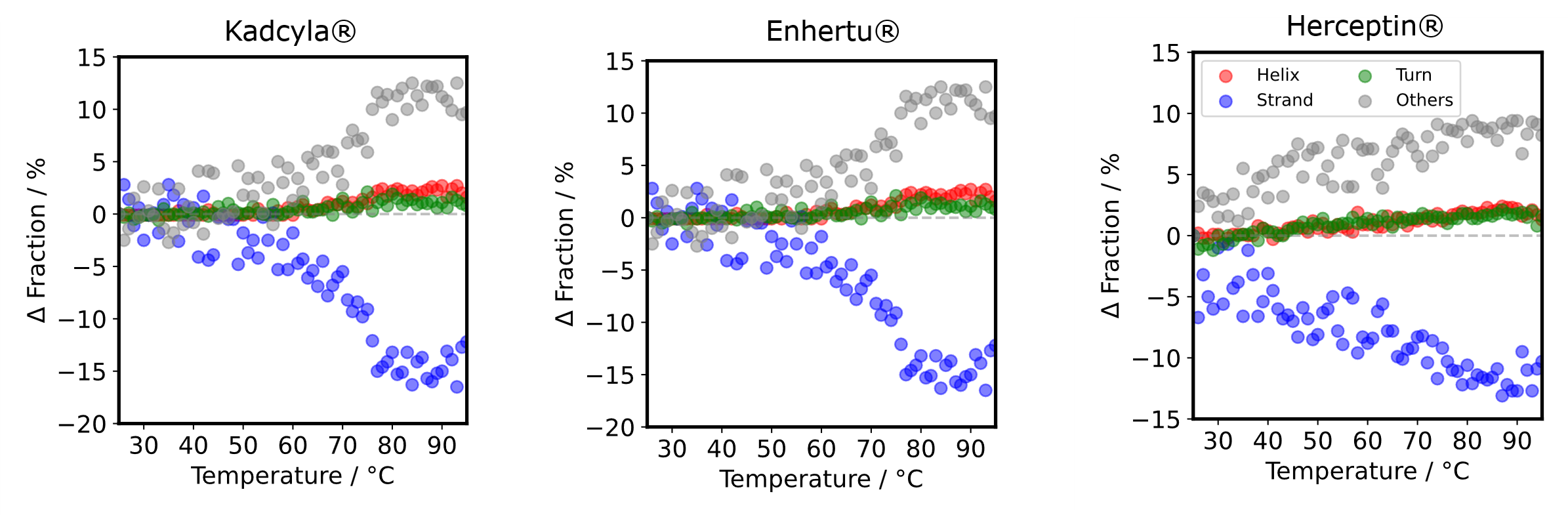
- Secondary structure compositions were estimated from the MRE spectra using BeStSel (https://bestsel.elte.hu)(2-5).
- lThe difference in fraction between 25 ºC and each temperature was plotted. As the temperature increases, the strand decreases and the others increases. The way how the Δ fraction changes is different in Herceptin® compared to Kadcyla® and Enhertu®.
References
Poster Session at PEGS Boston 2025 (The Essential Protein & Antibody Engineering Summit, May 12 – 16, 2025, in Boston, MA, USA)
Satoko Suzuki1,2, Ai Yamane1, Yasuo Horiguchi1, Taiji Oyama1, Hidetsugu Tabata3, Takehiko Wada2, András Micsonai4, József Kardos4, Ken-ichi Akao1
1JASCO Corporation, Hachioji, Tokyo 192-8537, Japan,
2Tohoku University, 2-1-1 Katahira, Aoba-ku, Sendai, Miyagi 980-8577, Japan,
3Teikyo University, 2-11-1 Kaga, Itabashi-ku, Tokyo 173-8605, Japan,
4ELTE NAP Neuroimmunology Research Group, Department of Biochemistry, Institute of Biology, ELTE Eötvös Loránd University, Budapest H-1117, Hungary
1.Saphire, E O et al. “Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design.” Science (New York, N.Y.) vol. 293,5532 (2001): 1155-9. doi:10.1126/science.1061692
2.Micsonai, András et al. “Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy.” Proceedings of the National Academy of Sciences of the United States of America vol. 112,24 (2015): E3095-103. doi:10.1073/pnas.1500851112
3.Micsonai, András et al. “BeStSel: a web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra.” Nucleic acids research vol. 46,W1 (2018): W315-W322. doi:10.1093/nar/gky497
4.Micsonai, András et al. “BeStSel: From Secondary Structure Analysis to Protein Fold Prediction by Circular Dichroism Spectroscopy.” Methods in molecular biology (Clifton, N.J.) vol. 2199 (2021): 175-189. doi:10.1007/978-1-0716-0892-0_11
5.Micsonai, András et al. “Disordered-Ordered Protein Binary Classification by Circular Dichroism Spectroscopy.” Frontiers in molecular biosciences vol. 9 863141. 3 May. 2022, doi:10.3389/fmolb.2022.863141
6.Micsonai, András et al. “BeStSel: webserver for secondary structure and fold prediction for protein CD spectroscopy.” Nucleic acids research vol. 50,W1 (2022): W90-W98. doi:10.1093/nar/gkac345