Introduction
Neurodegenerative disorders, including Alzheimer’s disease (AD) and Parkinson’s disease (PD), are characterized by progressive cognitive decline, and global efforts are underway to develop effective treatments. Key proteins such as β-amyloid, 1 a-synuclein, and tau are implicated in the pathogenesis of these diseases. the aggregation of β-amyloid plays a central role in neurodegenerative diseases such as AD, and research on this phenomenon is crucial for understanding the pathology and developing therapeutic strategies. β-amyloid exists extracellularly and undergoes an aggregation process, in which monomeric peptides form micellar intermediates under certain stimuli, which then further grow into fibrils (fibrous structures) or form amyloid plaques. These amyloid plaques accumulate between neurons, and ultimately lead to neuronal death. Understanding this aggregation process is essential for discovering new approaches to prevent or treat the disease. Circular dichroism (CD) spectroscopy is well known as a convenient and sensitive method for investigating the higher-order structure (HOS) of proteins in solution. This technique can monitor the changes in HOS and the formation of aggregates by measuring CD signals of β-amyloid peptide,2-4 making it valuable for elucidating disease mechanisms and advancing the development of effective therapeutics and preventive agents. In this presentation, we report the results of the evaluation of the performance of ionic liquids as inhibitors of amyloid aggregate formation and as solubilizers of β-amyloid peptide aggregates using CD spectroscopy and the BeStSel program, which allows accurate and detailed evaluation of secondary structure of proteins.
Experimental
Materials

Instrument and analysis program

Measurement method
- Sample conditions: 25 mM amyloid b-peptide dissolved in 67 mM phosphate buffer (pH 7.0) in the absence of IL or in the presence of 0.005% of three different ILs.
- Data acquisition: 400 mL sample was loaded into a 1 mm optical path length cell, and spectra were collected at 20 min intervals for 400 min.
- Measurement conditions: bandwidth 1 nm, scanning speed 50 nm / min, response 2 sec.
Results
Inhibition effect of ionic liquids
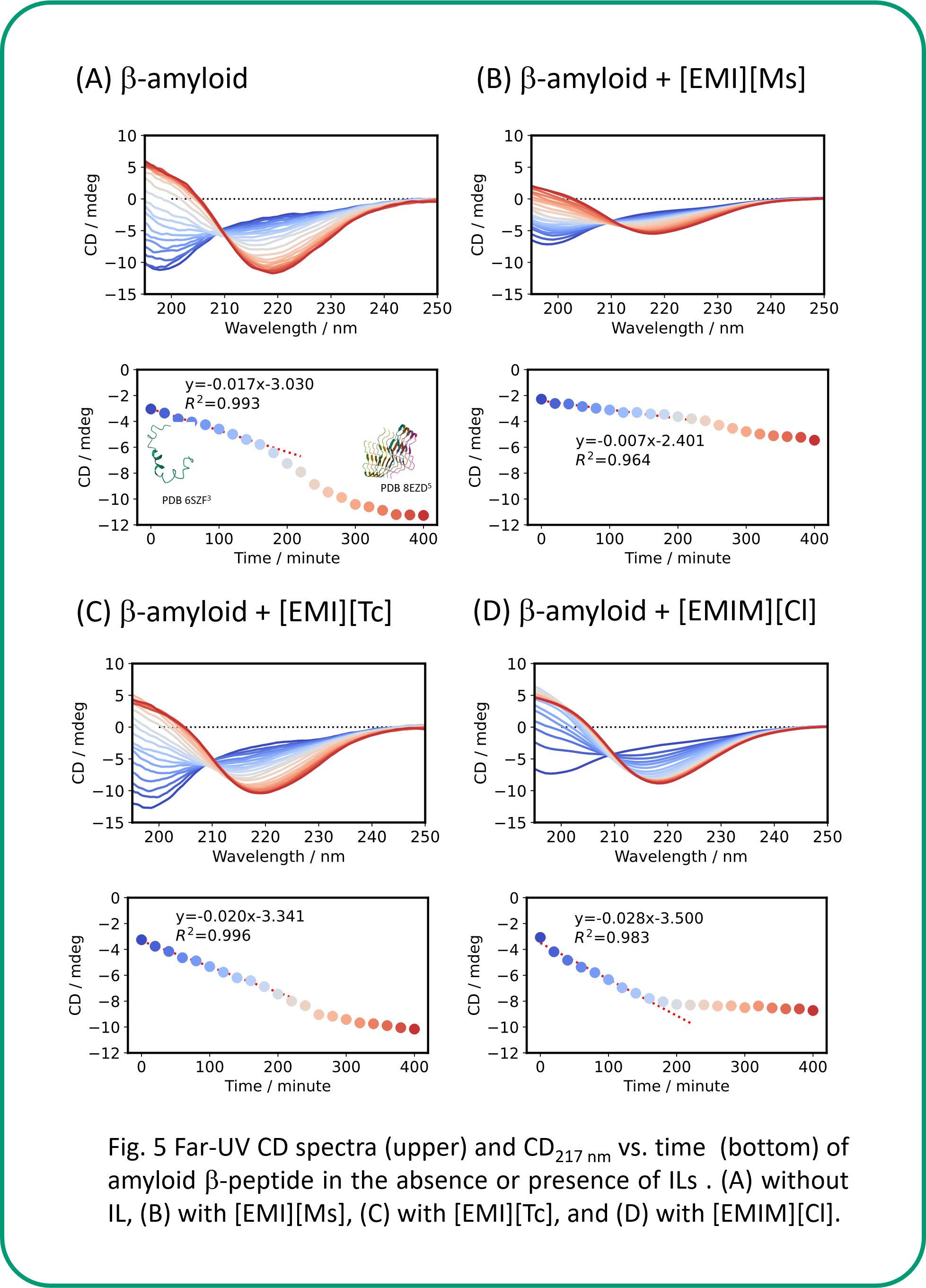
The CD signal at 217 nm derived from the b-sheet was decreased over time for amyloid β-peptide (Human, 1-42) due to aggregation (Fig. 5 A). In the presence of [EMI][Ms], the small change in the CD spectrum as well as the smaller initial amount of change in the CD signal just after the addition of IL than in the absence of IL (Fig. 5 B) indicate that IL inhibits the aggregation of peptides. On the other hand, for [EMI][Tc] and [EMIM][Cl], the initial change in CD signal just after the addition of IL was larger than that without IL, suggesting that IL promotes peptide aggregation in the first-step aggregate formation.
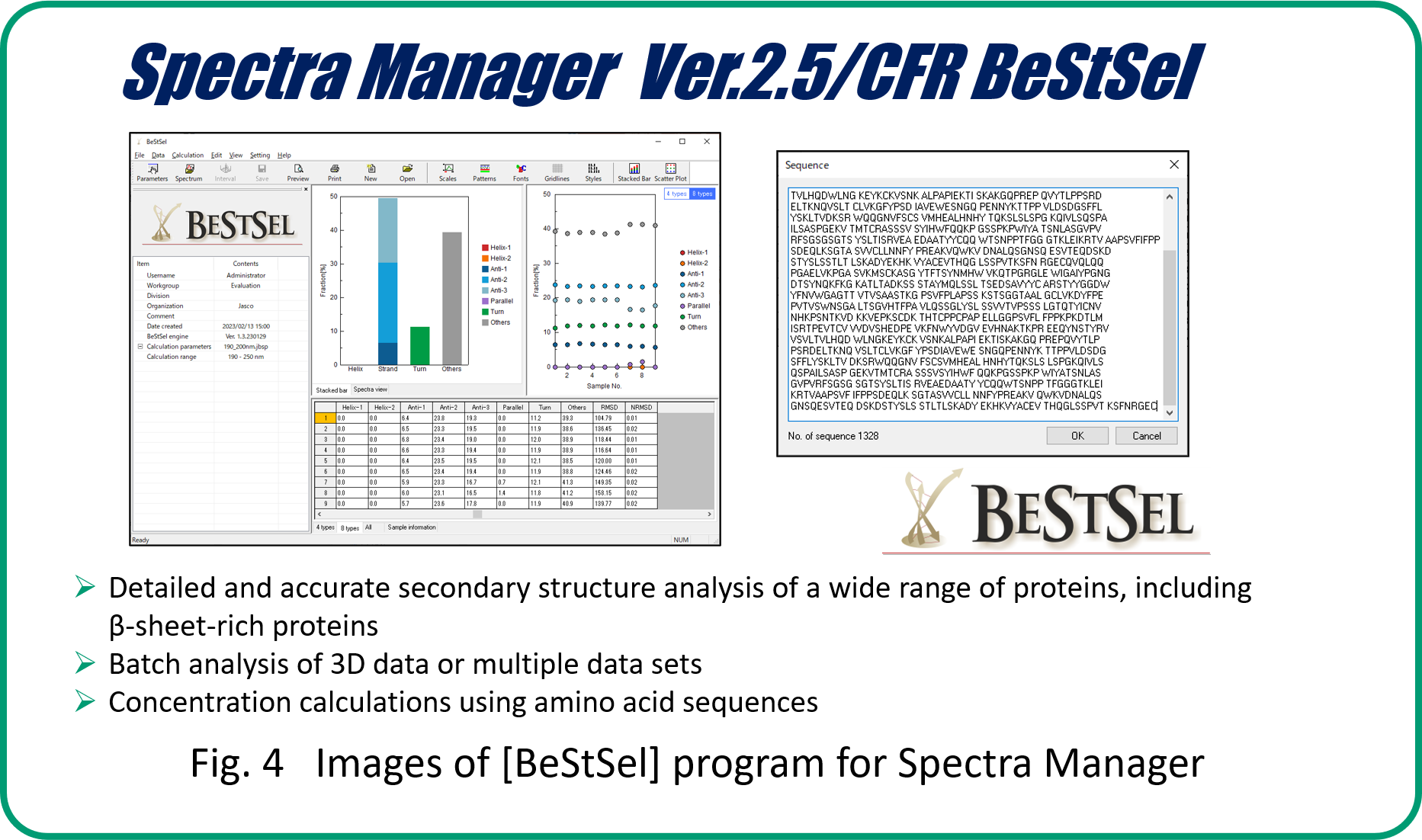
Secondary structure analysis using BeStSel
Secondary structure analysis using BeStSel allows us to evaluate the time dependence of the secondary structure fractions (Fig. 6 A-D). Overall, β-strands showed more or less an increasing trend, while others showed a decreasing trend. It is also possible to classify β-sheet into four types: parallel β-strand and three types of antiparallel β-strands.
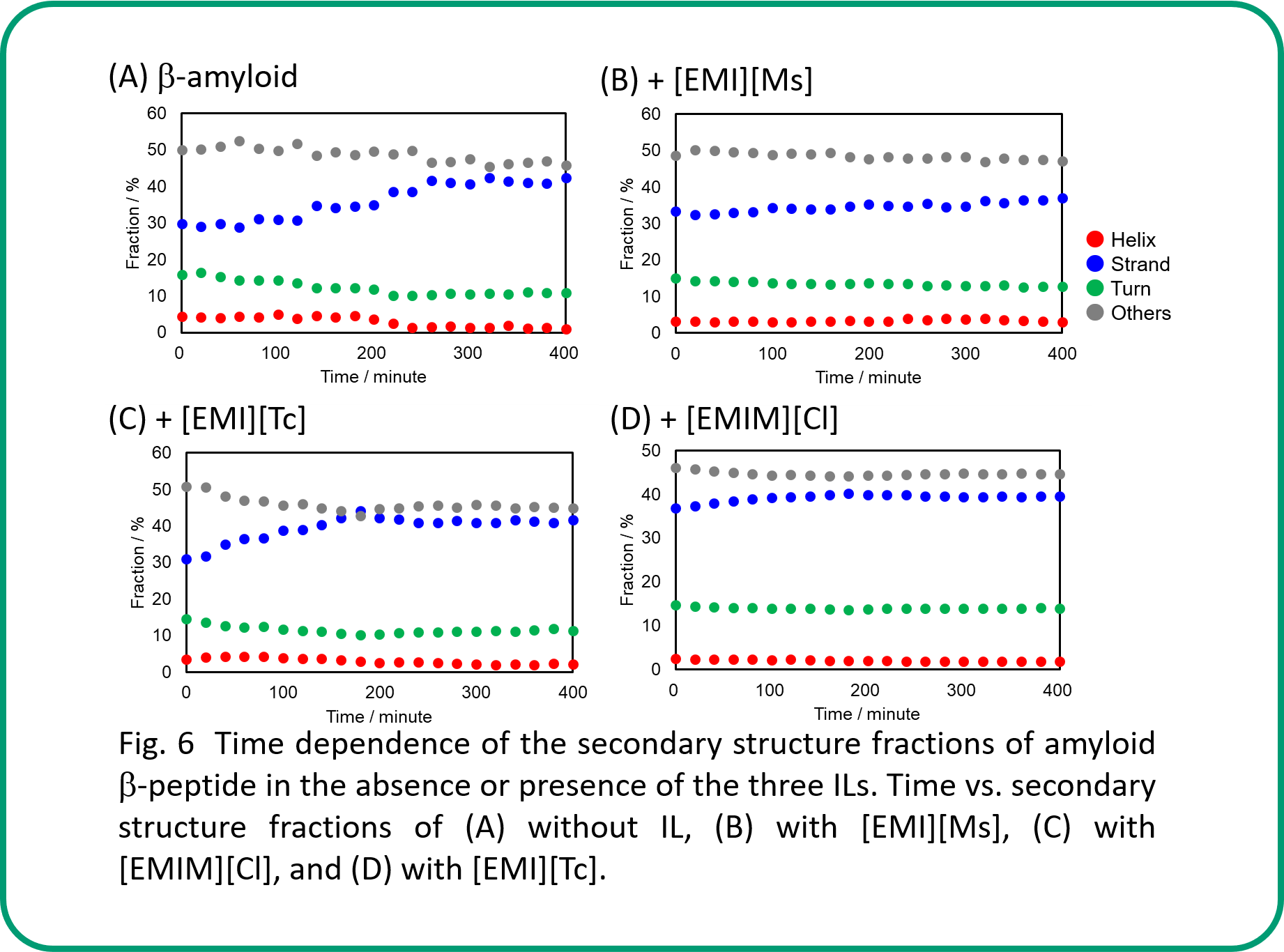
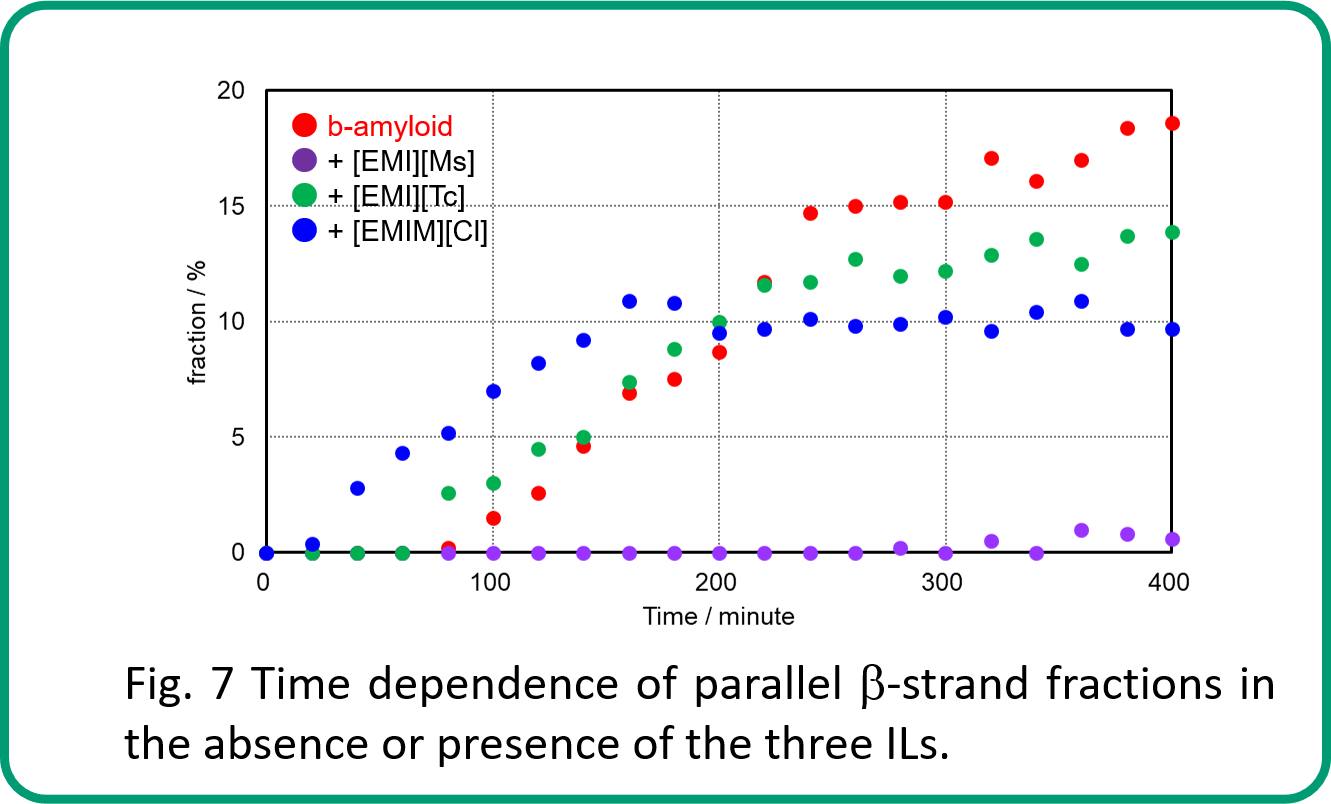
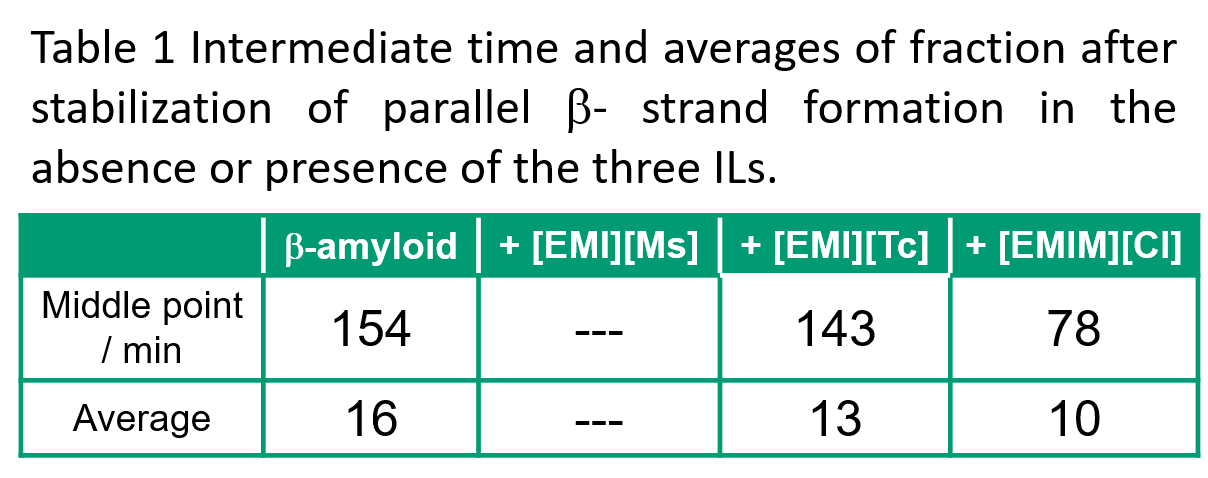
Fig. 7 shows the time dependence of the parallel β-strand fraction, which is especially important during aggregate formation. This result indicates that parallel β-strand is not formed when [EMI][Ms] is present. The intermediate time for the first stabilization of parallel β-strand formation and the average value of parallel β-strand fraction after this stabilization were as follows. Intermediate times were fastest in the presence of [EMIM][Cl], followed by [EMI][Tc], then β-amyloid only. The average was largest in the presence of β-amyloid only, followed by [EMI][Tc], then in the presence of [EMIM][Cl].
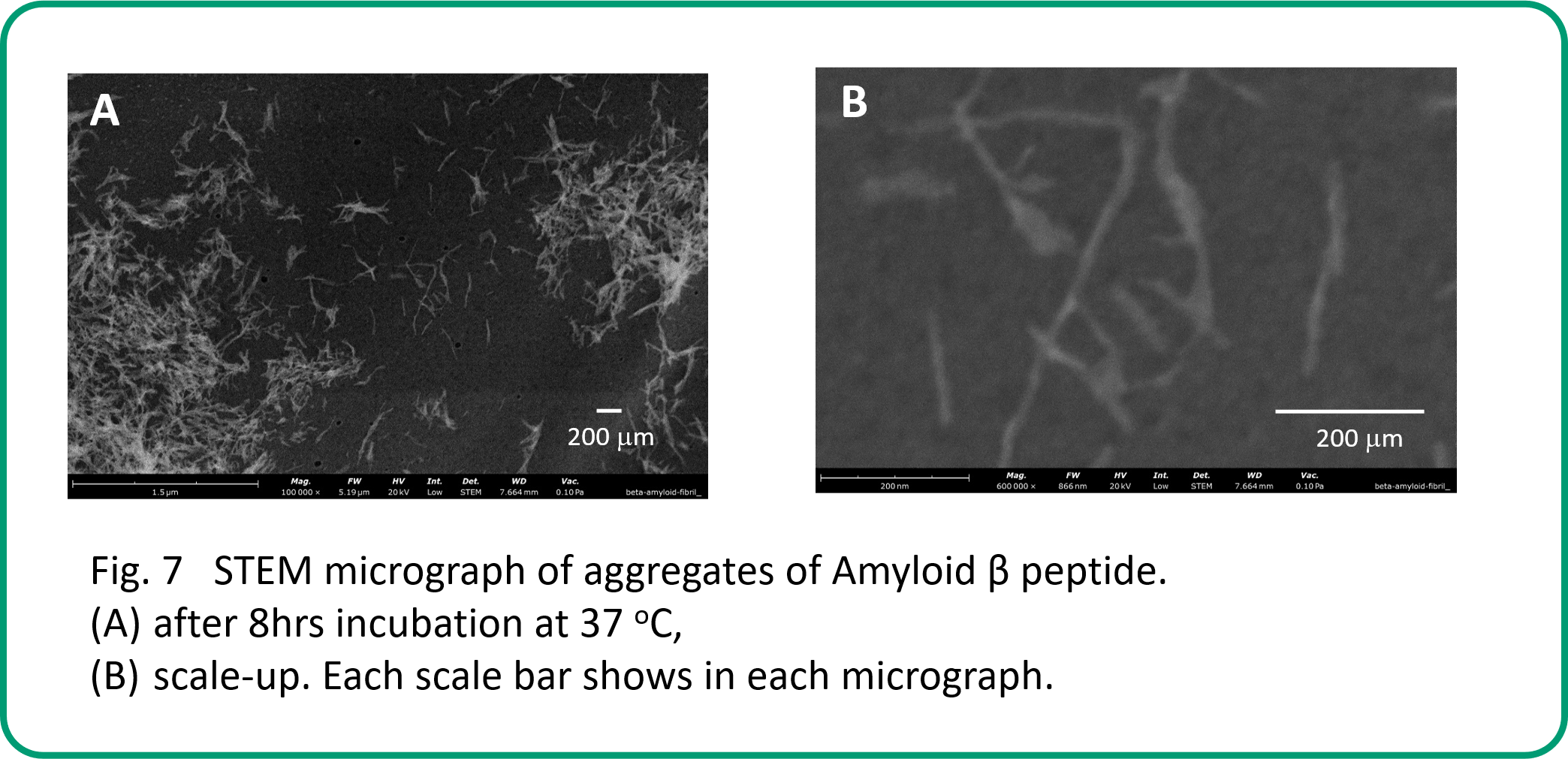
SEM analysis
Conclusion
- Only [EMI][Ms] ionic liquid inhibited the first-step aggregation of amyloid β-peptide (1-42).
- The results demonstrate that the combination of CD spectroscopy and the BestSel program is effective for the evaluation of the secondary structure fractions of amyloid β-peptides in detail.
References
1)J. Hardy, K. Duff, K. G. Hardy, J. Perez-Tur, M. Hutton (1998) Genetic dissection of Alzheimer‘s disease and related dementias: amyloid and its relationship to tau. Nature Neuroscience 1: 355-358.
2)(A) T. Takekiyo, Y. Yoshimura (2018) Suppression and dissolution of amyloid aggregates using ionic liquids. Biophysical Reviews 10: 853–860. (B) O. Oludayo, O. Afe, B. Strdel (2012) Structures of the Amyloid beta-Peptides A beta(1-40) and A beta(1-42) as Influenced by pH and a D-Peptide. The Journal of Physical Chemistry B 116: 3280-3291. (C) N. Debeljuh, C. J. Barrow, L. Henderson, N. Byrne (2011) Structure inducing ionic liquids—enhancement of alpha helicity in the Abeta(1–40) peptide from Alzheimer’s disease. Chemical Communications 47: 6371–6373.
3)Image from the RCSB PDB (rcsb.org) of PDB ID 6SZF (A. Santoro, M. Grimaldi, M. Buonocore, I. Stillitano, A. M D’Ursi (2021) Exploring the Early Stages of the Amyloid Aβ(1–42) Peptide Aggregation Process: An NMR Study. Pharmaceuticals 14: 732).
4)(A) A. Micsonai, F. Wien, L. Kernya, Y. H. Lee, Y. Goto, M. Réfrégiers, J. Kardos (2015) Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. PNAS 112: E3095-3103. (B) A. Micsonai, F. Wien, E. Bulyaki, J. Kun, E. Moussong, Y. H. Lee, Y. Goto, M. Réfrégiers, J. Kardos (2018) BeStSel: a web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra. Nucleic Acids Research 46: W315-W322. (C) A. Micsonai, E. Bulyaki, J. Kardos (2020) BeStSel: From Secondary Structure Analysis to Protein Fold Prediction by Circular Dichroism Spectroscopy. Methods in Molecular Biology 2199: 175-189. (D) A. Micsonai, E. Moussong, N. Murvai, A. Tantos, O. Toke, M. Réfrégiers, F. Wien, J. Kardos (2022) Disordered–Ordered Protein Binary Classification by Circular Dichroism Spectroscopy. Frontiers in Molecular Biosciences 9: 863141. (E) A. Micsonai, E. Moussong, F. Wien. E. Boros, H. Vadaszi, N. Murvai, Y. H Lee, T. Molnar, M. Réfrégiers, Y. Goto, A. Tantos. J. Kardos (2022) BeStSel: webserver for secondary structure and fold prediction for protein CD spectroscopy. Nucleic Acids Research 50: W90-W98.
5)Image from the RCSB PDB (rcsb.org) of PDB ID 8EZD (M. Lee, W. M. Yau, J. M. Louis, R. Tycko (2023) Structures of brain-derived 42-residue amyloid-β fibril polymorphs with unusual molecular conformations and intermolecular interactions. Biophysics and computational biology 120: e2218831120).






